Input data and parameters
QualiMap command line
| qualimap bamqc -bam /data/dnb03/galaxy_db/files/5/d/d/dataset_5ddca281-f41f-47de-9e25-a82d3cc29497.dat -c -nw 400 -hm 3 -sd |
Alignment
| Command line: | bwa mem -t 8 -v 1 -R @RG\tID:Cutadapt_on_data_12_and_data_11__Read_\tSM:Cutadapt_on_data_12_and_data_11__Read_\tPL:ILLUMINA\tLB:Cutadapt_on_data_12_and_data_11__Read_\tPU:run /data/db/data_managers/hg19/bwa_mem_index/hg19/hg19.fa /data/dnb03/galaxy_db/files/f/6/7/dataset_f67c75a7-7d88-4dc5-be19-3b548e341400.dat /data/dnb03/galaxy_db/files/c/7/9/dataset_c799528a-8dcb-4e84-8648-3c5da85bce91.dat |
| Draw chromosome limits: | yes |
| Analyze overlapping paired-end reads: | yes |
| Program: | bwa (0.7.17-r1188) |
| Analysis date: | Sat May 22 06:50:45 CEST 2021 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | yes (only flagged) |
| Number of windows: | 400 |
| BAM file: | /data/dnb03/galaxy_db/files/5/d/d/dataset_5ddca281-f41f-47de-9e25-a82d3cc29497.dat |
Summary
Warnings
| No flagged duplicates are detected | Make sure duplicate alignments are flagged in the BAM file or apply a different skip duplicates mode. |
Globals
| Reference size | 3,137,161,264 |
| Number of reads | 63,229,532 |
| Mapped reads | 63,193,842 / 99.94% |
| Unmapped reads | 35,690 / 0.06% |
| Mapped paired reads | 63,193,842 / 99.94% |
| Mapped reads, first in pair | 31,603,270 / 49.98% |
| Mapped reads, second in pair | 31,590,572 / 49.96% |
| Mapped reads, both in pair | 63,167,936 / 99.9% |
| Mapped reads, singletons | 25,906 / 0.04% |
| Secondary alignments | 0 |
| Supplementary alignments | 374,257 / 0.59% |
| Read min/max/mean length | 20 / 101 / 98.7 |
| Overlapping read pairs | 8,762,868 / 27.72% |
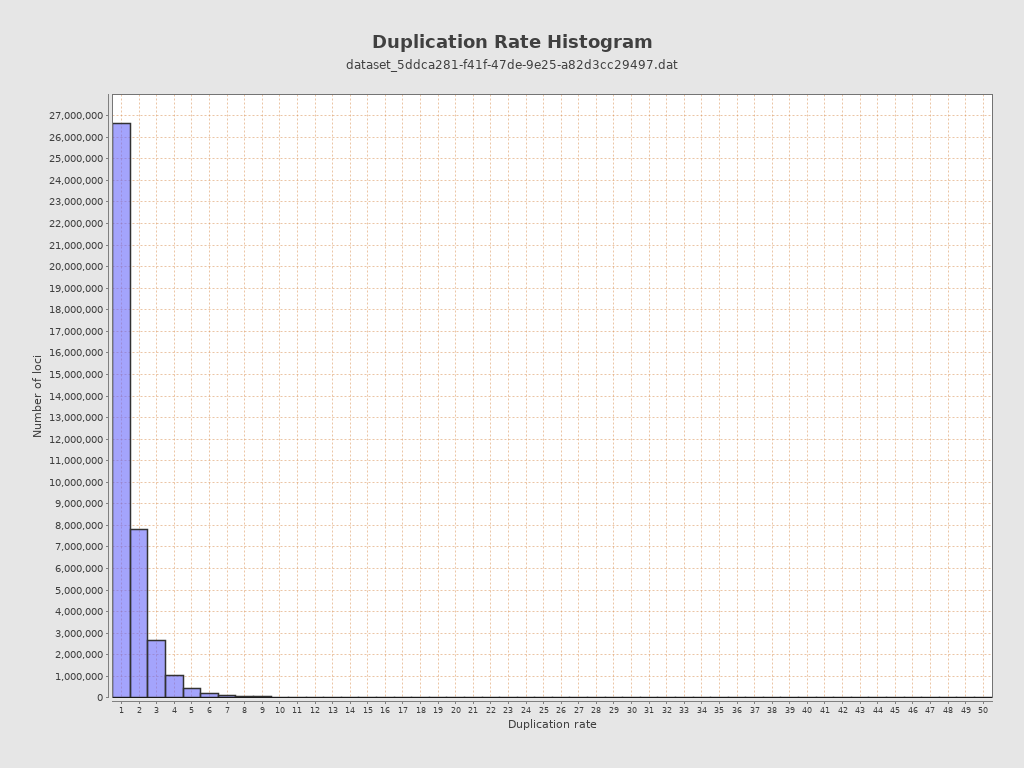
| Duplicated reads (flagged) | 0 / 0% |
| Duplicated reads (estimated) | 24,326,393 / 38.47% |
| Duplication rate | 32.13% |
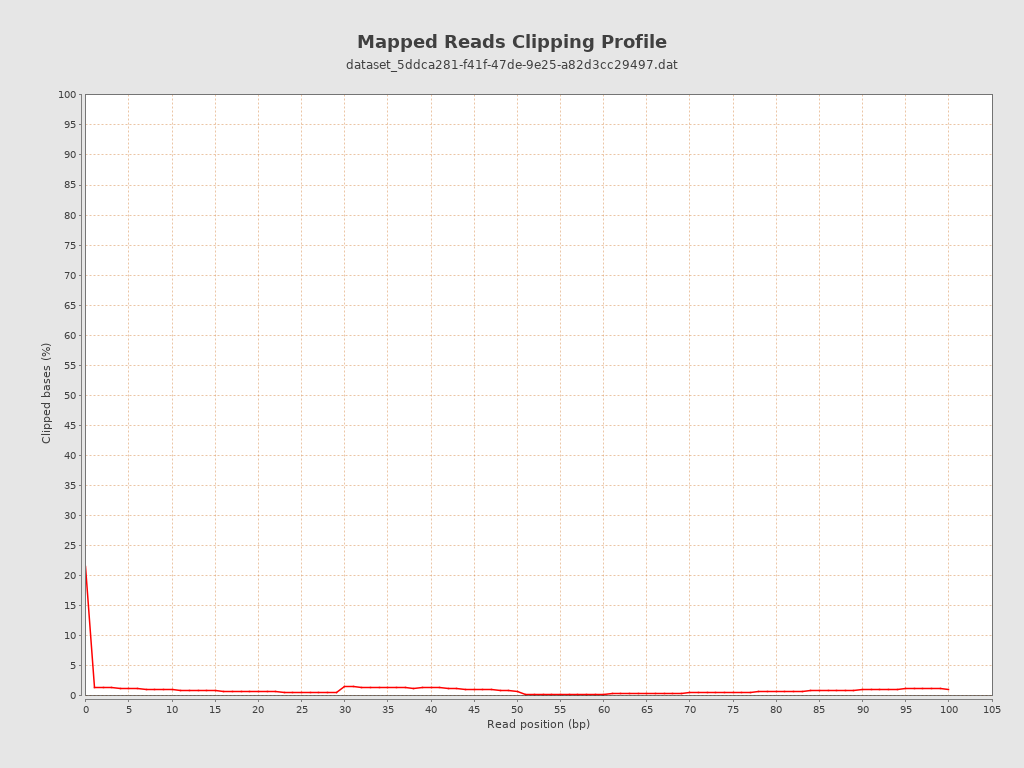
| Clipped reads | 1,874,815 / 2.97% |
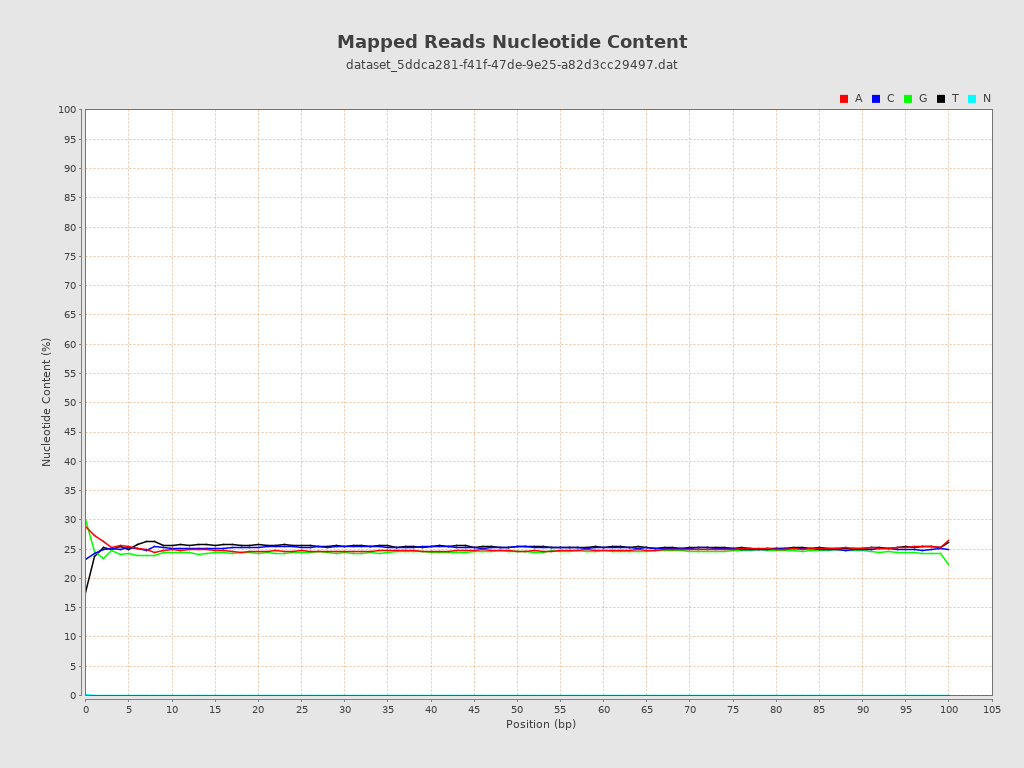
ACGT Content
| Number/percentage of A's | 1,548,501,483 / 24.96% |
| Number/percentage of C's | 1,561,248,924 / 25.16% |
| Number/percentage of T's | 1,571,612,057 / 25.33% |
| Number/percentage of G's | 1,523,328,025 / 24.55% |
| Number/percentage of N's | 179,883 / 0% |
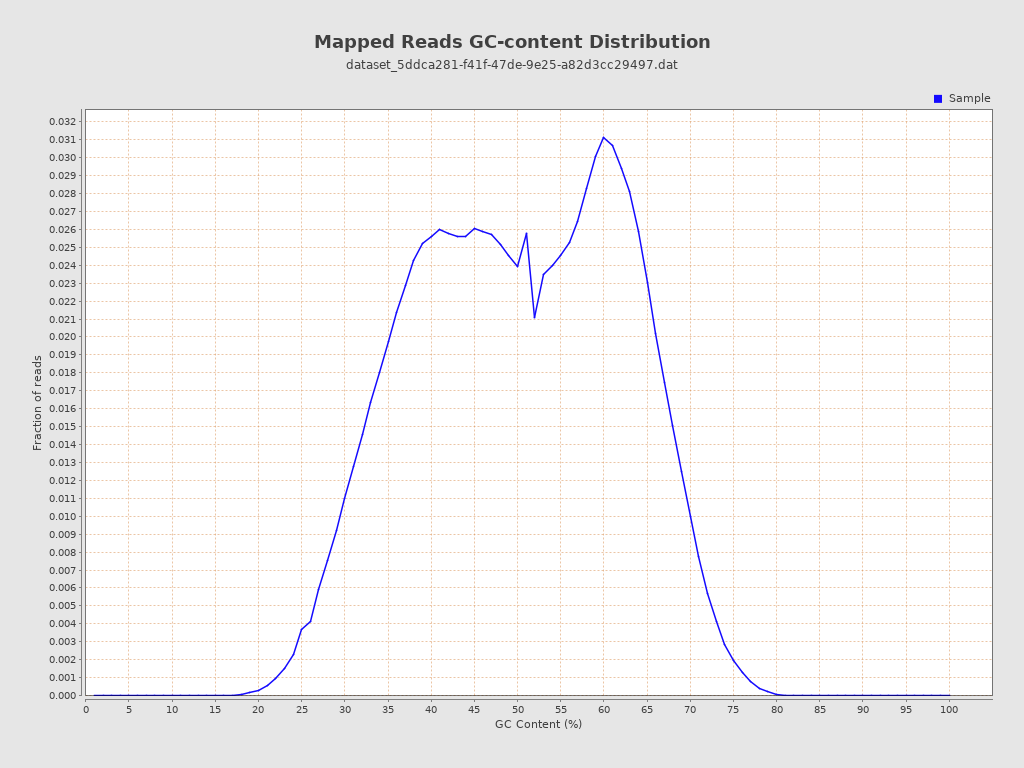
| GC Percentage | 49.71% |
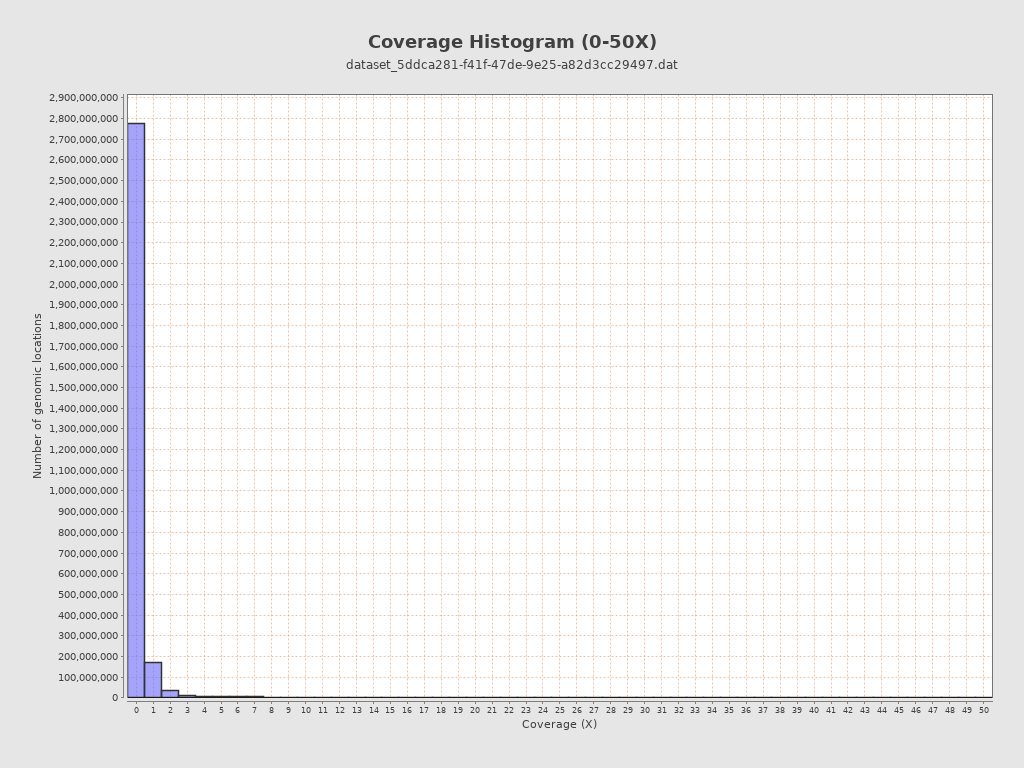
Coverage
| Mean | 1.9781 |
| Standard Deviation | 20.9942 |
| Mean (paired-end reads overlap ignored) | 1.89 |
Mapping Quality
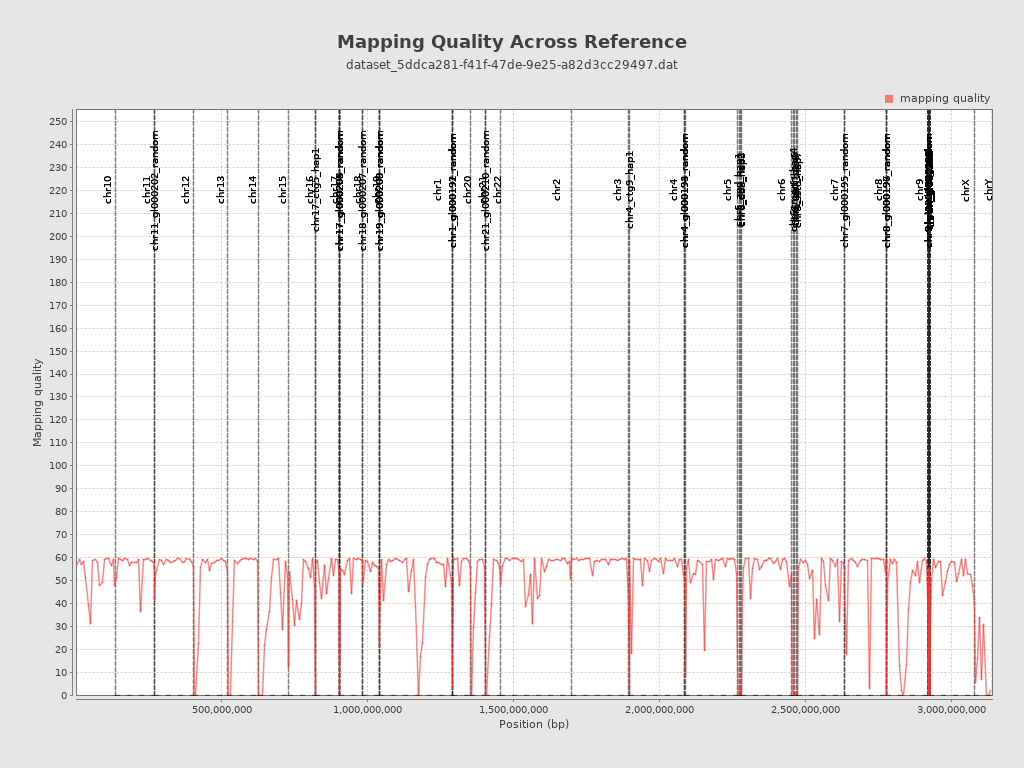
| Mean Mapping Quality | 46.54 |
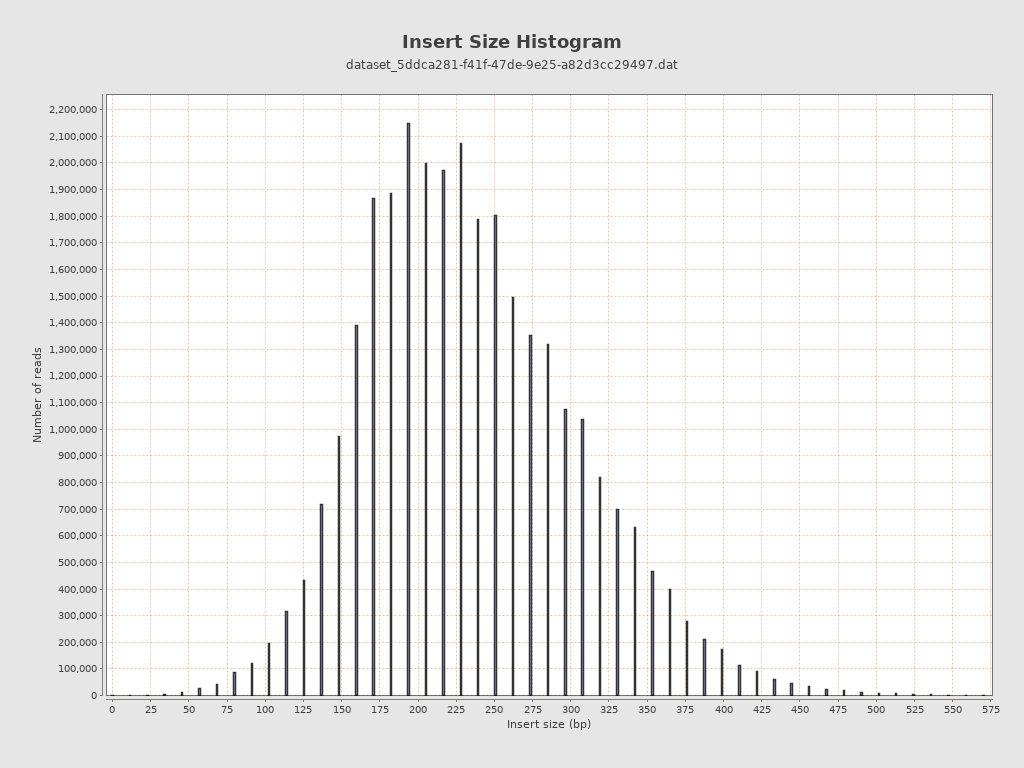
Insert size
| Mean | 148,760.09 |
| Standard Deviation | 3,800,455.14 |
| P25/Median/P75 | 191 / 233 / 285 |
Mismatches and indels
| General error rate | 0.23% |
| Mismatches | 13,771,546 |
| Insertions | 206,736 |
| Mapped reads with at least one insertion | 0.32% |
| Deletions | 302,357 |
| Mapped reads with at least one deletion | 0.47% |
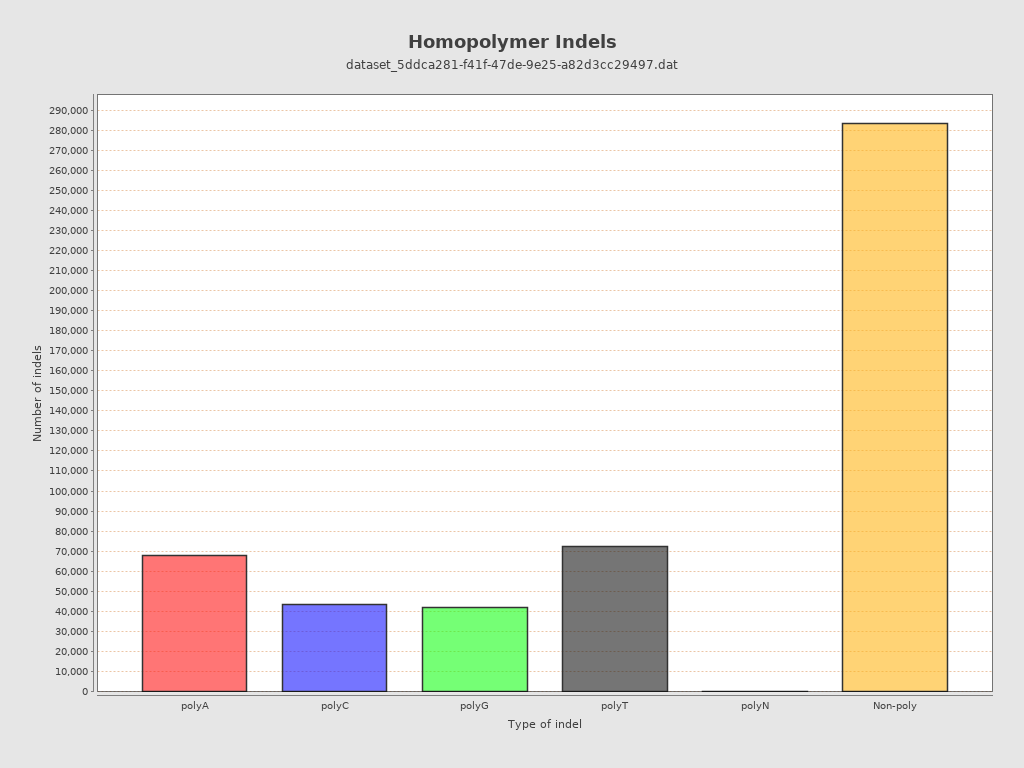
| Homopolymer indels | 44.34% |
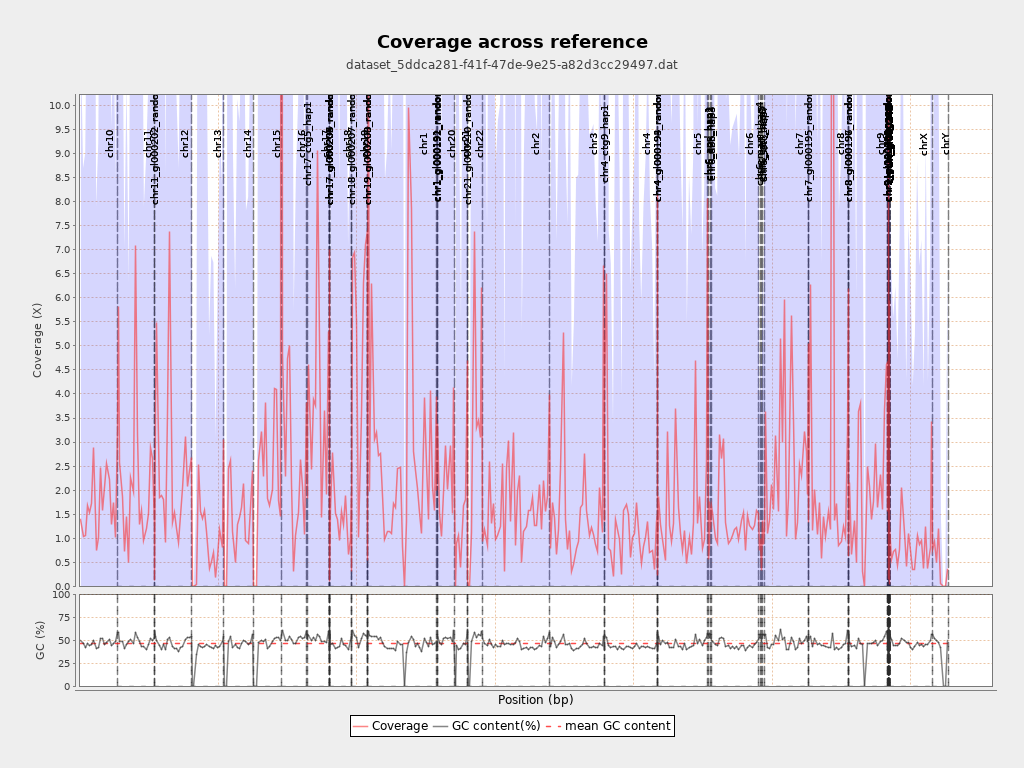
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| chr10 | 135534747 | 226459672 | 1.6709 | 11.7559 |
| chr11 | 135006516 | 301417296 | 2.2326 | 13.7836 |
| chr11_gl000202_random | 40103 | 4504 | 0.1123 | 0.3602 |
| chr12 | 133851895 | 332192260 | 2.4818 | 14.6911 |
| chr13 | 115169878 | 104993007 | 0.9116 | 8.0278 |
| chr14 | 107349540 | 148440162 | 1.3828 | 11.4751 |
| chr15 | 102531392 | 230398555 | 2.2471 | 14.9382 |
| chr16 | 90354753 | 232681440 | 2.5752 | 15.2916 |
| chr17_ctg5_hap1 | 1680828 | 3067487 | 1.825 | 9.582 |
| chr17 | 81195210 | 332216528 | 4.0916 | 21.735 |
| chr17_gl000203_random | 37498 | 3836 | 0.1023 | 0.3278 |
| chr17_gl000204_random | 81310 | 22807 | 0.2805 | 2.3357 |
| chr17_gl000205_random | 174588 | 1222201 | 7.0005 | 65.0947 |
| chr17_gl000206_random | 41001 | 4977 | 0.1214 | 0.4971 |
| chr18 | 78077248 | 114289167 | 1.4638 | 12.1719 |
| chr18_gl000207_random | 4262 | 1291 | 0.3029 | 0.7327 |
| chr19 | 59128983 | 324445027 | 5.4871 | 22.6243 |
| chr19_gl000208_random | 92689 | 49514 | 0.5342 | 1.7959 |
| chr19_gl000209_random | 159169 | 738663 | 4.6407 | 13.8984 |
| chr1 | 249250621 | 670725376 | 2.691 | 19.7168 |
| chr1_gl000191_random | 106433 | 211884 | 1.9908 | 8.0852 |
| chr1_gl000192_random | 547496 | 1435490 | 2.6219 | 11.3895 |
| chr20 | 63025520 | 139397564 | 2.2118 | 13.6235 |
| chr21 | 48129895 | 71677910 | 1.4893 | 13.5691 |
| chr21_gl000210_random | 27682 | 3729 | 0.1347 | 0.5361 |
| chr22 | 51304566 | 156088612 | 3.0424 | 19.6788 |
| chr2 | 243199373 | 392236321 | 1.6128 | 11.8323 |
| chr3 | 198022430 | 300328322 | 1.5166 | 12.5788 |
| chr4_ctg9_hap1 | 590426 | 790984 | 1.3397 | 8.3684 |
| chr4 | 191154276 | 239632775 | 1.2536 | 18.4492 |
| chr4_gl000193_random | 189789 | 285875 | 1.5063 | 20.0663 |
| chr4_gl000194_random | 191469 | 1249904 | 6.528 | 69.6571 |
| chr5 | 180915260 | 282159979 | 1.5596 | 15.8471 |
| chr6_apd_hap1 | 4622290 | 2536529 | 0.5488 | 4.3881 |
| chr6_cox_hap2 | 4795371 | 6055597 | 1.2628 | 5.5068 |
| chr6_dbb_hap3 | 4610396 | 5434598 | 1.1788 | 5.4373 |
| chr6 | 171115067 | 246975243 | 1.4433 | 10.4851 |
| chr6_mann_hap4 | 4683263 | 3533811 | 0.7546 | 3.1982 |
| chr6_mcf_hap5 | 4833398 | 4838390 | 1.001 | 4.2553 |
| chr6_qbl_hap6 | 4611984 | 5448977 | 1.1815 | 5.2069 |
| chr6_ssto_hap7 | 4928567 | 4544457 | 0.9221 | 4.3589 |
| chr7 | 159138663 | 424157631 | 2.6653 | 30.476 |
| chr7_gl000195_random | 182896 | 922710 | 5.045 | 45.7039 |
| chr8 | 146364022 | 406320336 | 2.7761 | 64.6299 |
| chr8_gl000196_random | 38914 | 24510 | 0.6299 | 3.2829 |
| chr8_gl000197_random | 37175 | 24007 | 0.6458 | 3.3159 |
| chr9 | 141213431 | 291703661 | 2.0657 | 15.6952 |
| chr9_gl000198_random | 90085 | 139964 | 1.5537 | 14.8952 |
| chr9_gl000199_random | 169874 | 576082 | 3.3912 | 9.2308 |
| chr9_gl000200_random | 187035 | 147953 | 0.791 | 5.9327 |
| chr9_gl000201_random | 36148 | 106616 | 2.9494 | 7.7289 |
| chrM | 16571 | 630673 | 38.0588 | 12.0505 |
| chrUn_gl000211 | 166566 | 2134510 | 12.8148 | 36.5888 |
| chrUn_gl000212 | 186858 | 4538023 | 24.2859 | 112.1768 |
| chrUn_gl000213 | 164239 | 964998 | 5.8756 | 37.2243 |
| chrUn_gl000214 | 137718 | 4397414 | 31.9306 | 109.4624 |
| chrUn_gl000215 | 172545 | 429017 | 2.4864 | 13.6206 |
| chrUn_gl000216 | 172294 | 352142 | 2.0438 | 10.3334 |
| chrUn_gl000217 | 172149 | 283459 | 1.6466 | 14.3224 |
| chrUn_gl000218 | 161147 | 899406 | 5.5813 | 33.8379 |
| chrUn_gl000219 | 179198 | 1068731 | 5.964 | 51.5973 |
| chrUn_gl000220 | 161802 | 6534819 | 40.3878 | 295.9962 |
| chrUn_gl000221 | 155397 | 3185408 | 20.4985 | 92.0217 |
| chrUn_gl000222 | 186861 | 971433 | 5.1987 | 38.1596 |
| chrUn_gl000223 | 180455 | 416917 | 2.3104 | 8.3677 |
| chrUn_gl000224 | 179693 | 549678 | 3.059 | 17.9514 |
| chrUn_gl000225 | 211173 | 890103 | 4.215 | 29.9661 |
| chrUn_gl000226 | 15008 | 485537 | 32.3519 | 23.8515 |
| chrUn_gl000227 | 128374 | 488740 | 3.8072 | 21.4372 |
| chrUn_gl000228 | 129120 | 890296 | 6.8951 | 26.721 |
| chrUn_gl000229 | 19913 | 341463 | 17.1477 | 45.9831 |
| chrUn_gl000230 | 43691 | 316003 | 7.2327 | 26.3145 |
| chrUn_gl000231 | 27386 | 279599 | 10.2096 | 40.4918 |
| chrUn_gl000232 | 40652 | 95558 | 2.3506 | 19.4664 |
| chrUn_gl000233 | 45941 | 6274 | 0.1366 | 0.3941 |
| chrUn_gl000234 | 40531 | 123923 | 3.0575 | 27.4165 |
| chrUn_gl000235 | 34474 | 51390 | 1.4907 | 10.6756 |
| chrUn_gl000236 | 41934 | 143610 | 3.4247 | 15.2462 |
| chrUn_gl000237 | 45867 | 176030 | 3.8378 | 26.9727 |
| chrUn_gl000238 | 39939 | 293602 | 7.3513 | 21.8244 |
| chrUn_gl000239 | 33824 | 57281 | 1.6935 | 7.5674 |
| chrUn_gl000240 | 41933 | 50770 | 1.2107 | 6.6023 |
| chrUn_gl000241 | 42152 | 250185 | 5.9353 | 22.4816 |
| chrUn_gl000242 | 43523 | 156783 | 3.6023 | 11.3843 |
| chrUn_gl000243 | 43341 | 83068 | 1.9166 | 12.5309 |
| chrUn_gl000244 | 39929 | 194502 | 4.8712 | 28.9173 |
| chrUn_gl000245 | 36651 | 7242 | 0.1976 | 0.4849 |
| chrUn_gl000246 | 38154 | 45943 | 1.2041 | 7.5211 |
| chrUn_gl000247 | 36422 | 9329 | 0.2561 | 1.3717 |
| chrUn_gl000248 | 39786 | 1304 | 0.0328 | 0.1819 |
| chrUn_gl000249 | 38502 | 63238 | 1.6425 | 6.5337 |
| chrX | 155270560 | 132966699 | 0.8564 | 6.5504 |
| chrY | 59373566 | 27363160 | 0.4609 | 7.3415 |