![[OK]](tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | SRR8836584__fastq-dump__uncompressed |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 14394884 |
| Filtered Sequences | 0 |
| Sequence length | 126 |
| %GC | 35 |
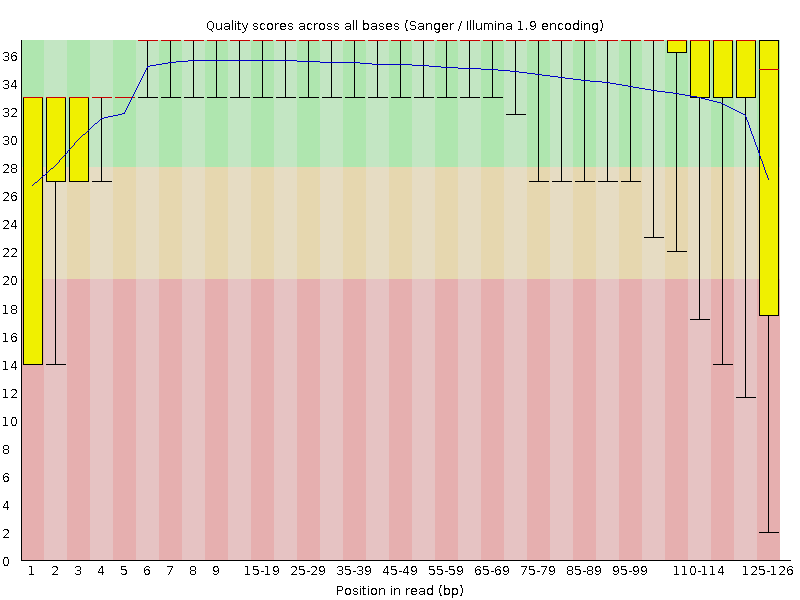
![[OK]](tick.png) Per base sequence quality
Per base sequence quality
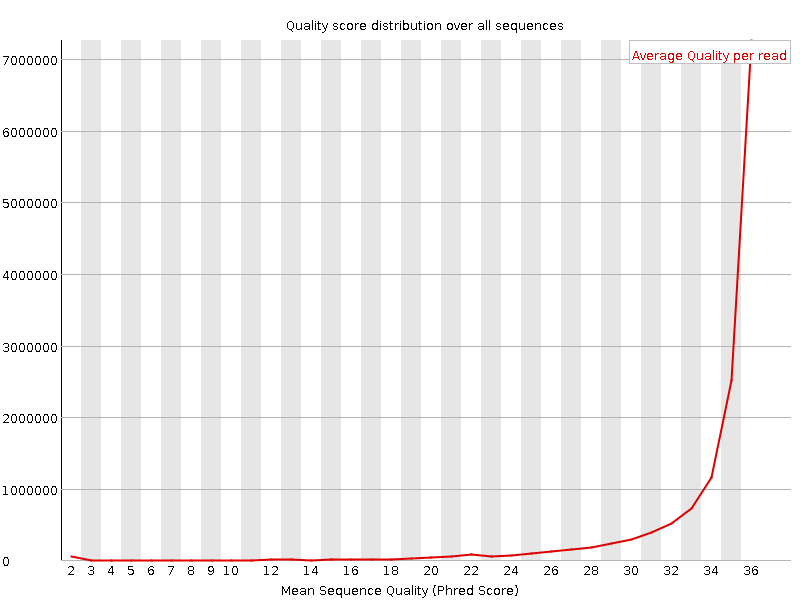
![[OK]](tick.png) Per sequence quality scores
Per sequence quality scores
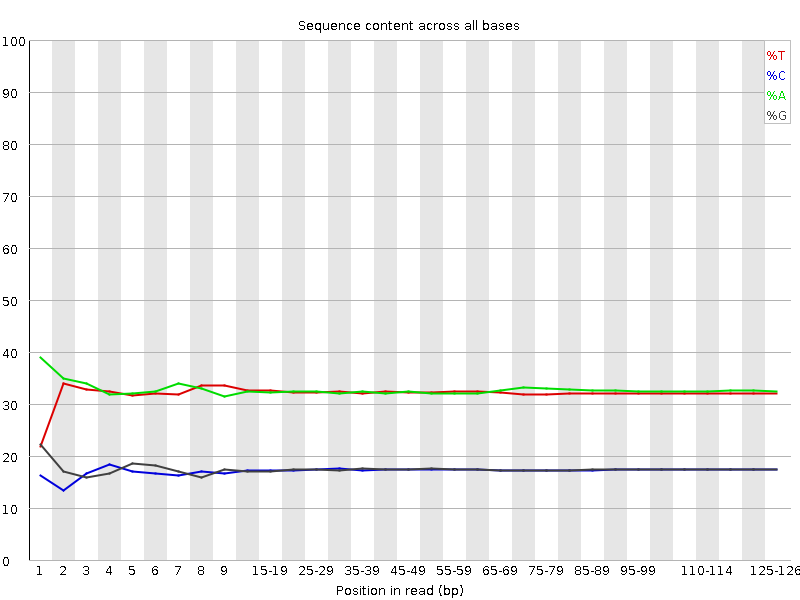
![[FAIL]](error.png) Per base sequence content
Per base sequence content
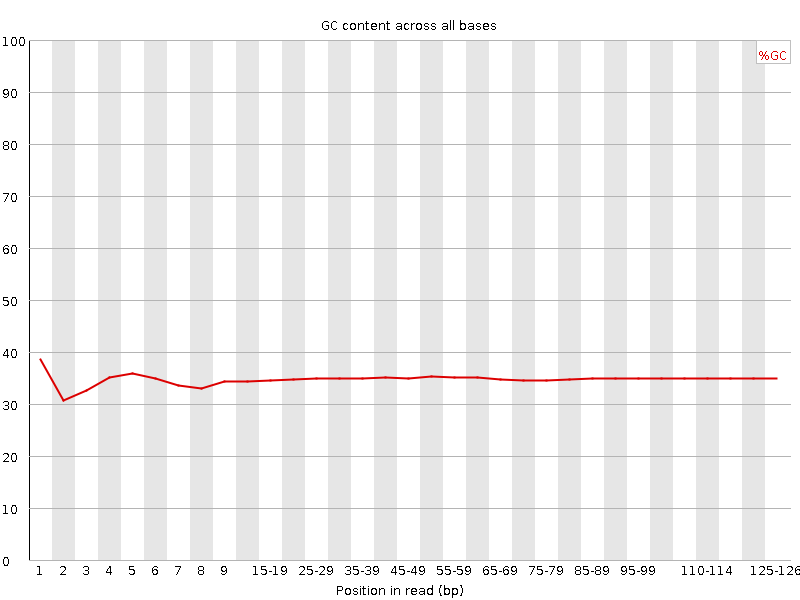
![[OK]](tick.png) Per base GC content
Per base GC content
![[OK]](tick.png) Per sequence GC content
Per sequence GC content
![[OK]](tick.png) Per base N content
Per base N content
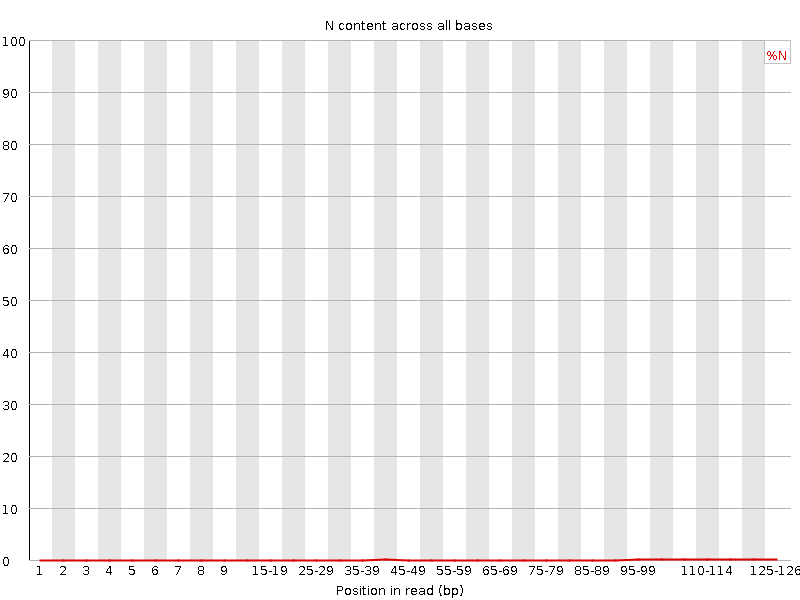
![[OK]](tick.png) Sequence Length Distribution
Sequence Length Distribution
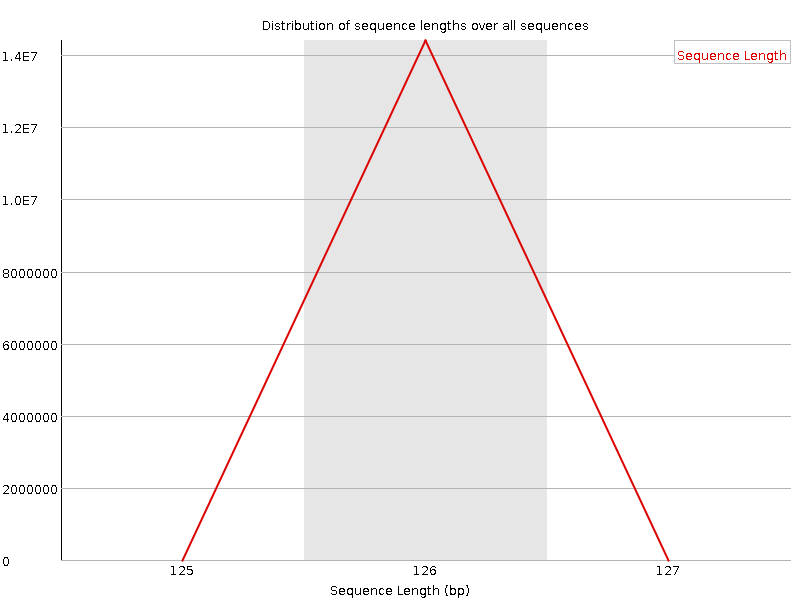
![[FAIL]](error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACCTGAAGCTATCTCGTAT | 73693 | 0.5119388249325246 | TruSeq Adapter, Index 7 (97% over 37bp) |
| GATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTTCAGAGCCGTGTAGATCT | 60466 | 0.4200520129234804 | Illumina Single End PCR Primer 1 (96% over 32bp) |
![[FAIL]](error.png) Kmer Content
Kmer Content
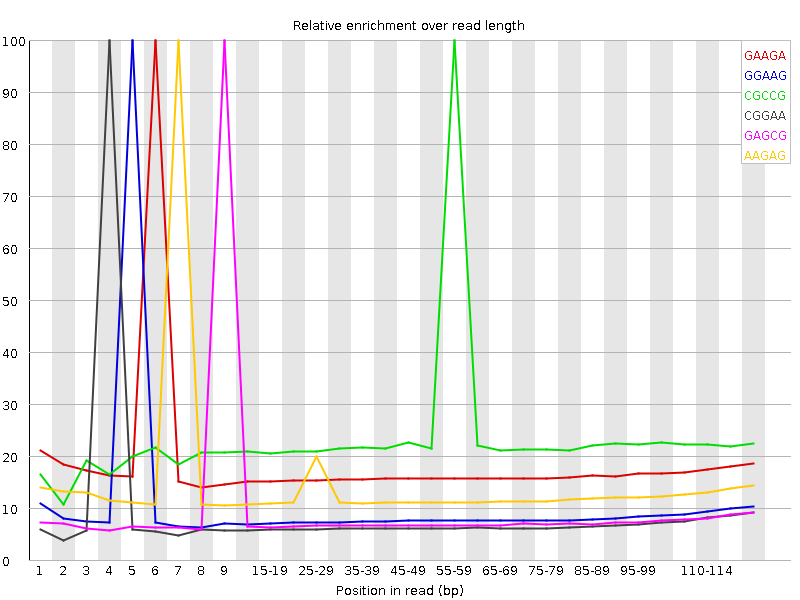
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GAAGA | 3974220 | 2.111342 | 12.467978 | 6 |
| GGAAG | 1896585 | 1.8772246 | 21.358154 | 5 |
| CGCCG | 451880 | 1.574039 | 6.354757 | 55-59 |
| CGGAA | 1565040 | 1.556183 | 21.11171 | 4 |
| GAGCG | 839960 | 1.5560771 | 19.651642 | 9 |
| AAGAG | 2868595 | 1.523968 | 11.824492 | 7 |
| AGAGC | 1483655 | 1.4752586 | 20.860527 | 8 |
| GAGCA | 1407375 | 1.3994104 | 11.113014 | 9 |
| ATCGG | 1234550 | 1.2411278 | 20.972294 | 2 |
| TCGGA | 1092695 | 1.0985171 | 20.970297 | 3 |
| GATCG | 967100 | 0.97225285 | 20.959332 | 1 |