Input data and parameters
QualiMap command line
| qualimap bamqc -bam /data/dnb03/galaxy_db/files/8/f/f/dataset_8ff305d9-a6f2-468b-baad-87cda8731209.dat -c -nw 400 -hm 3 -sd |
Alignment
| Command line: | bwa mem -t 8 -v 1 -R @RG\tID:SRR2153872_\tSM:SRR2153872_\tPL:ILLUMINA\tLB:SRR2153872_\tPU:run /data/db/data_managers/hg19/bwa_mem_index/hg19/hg19.fa /data/dnb03/galaxy_db/files/4/8/3/dataset_483e344f-5ee6-4bf7-9596-f2738fac16b7.dat /data/dnb03/galaxy_db/files/1/e/3/dataset_1e325183-ad98-443c-aa31-f5879990317b.dat |
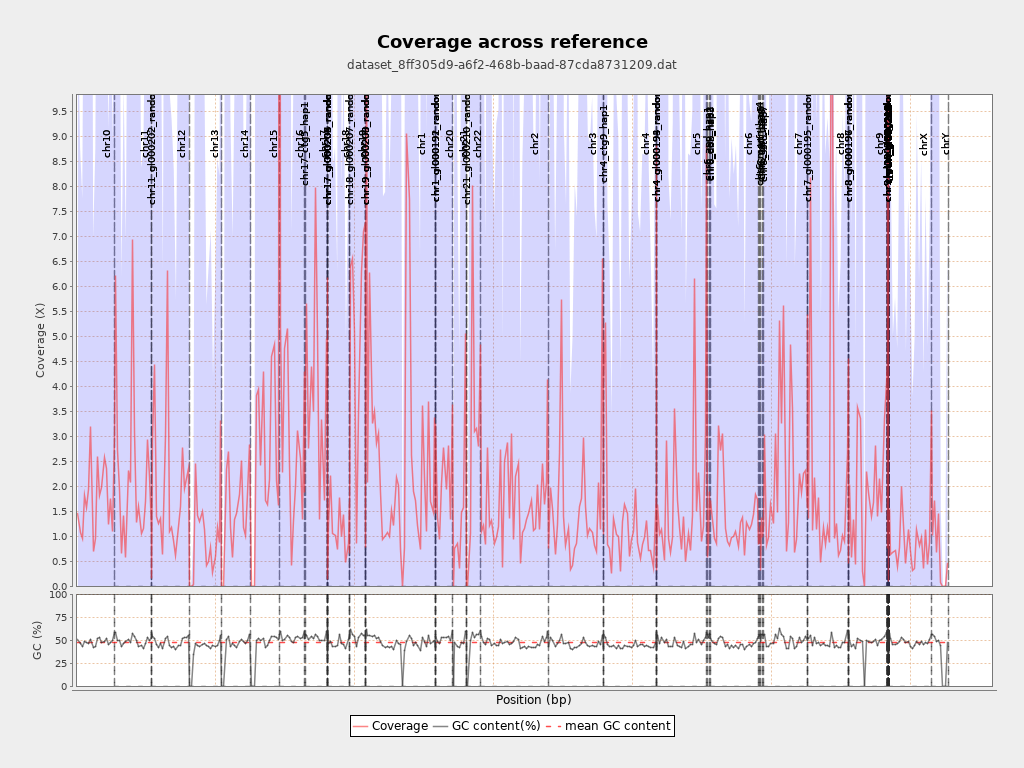
| Draw chromosome limits: | yes |
| Analyze overlapping paired-end reads: | yes |
| Program: | bwa (0.7.17-r1188) |
| Analysis date: | Sat May 22 06:49:29 CEST 2021 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | yes (only flagged) |
| Number of windows: | 400 |
| BAM file: | /data/dnb03/galaxy_db/files/8/f/f/dataset_8ff305d9-a6f2-468b-baad-87cda8731209.dat |
Summary
Warnings
| No flagged duplicates are detected | Make sure duplicate alignments are flagged in the BAM file or apply a different skip duplicates mode. |
Globals
| Reference size | 3,137,161,264 |
| Number of reads | 60,078,754 |
| Mapped reads | 59,779,306 / 99.5% |
| Unmapped reads | 299,448 / 0.5% |
| Mapped paired reads | 59,779,306 / 99.5% |
| Mapped reads, first in pair | 29,778,330 / 49.57% |
| Mapped reads, second in pair | 30,000,976 / 49.94% |
| Mapped reads, both in pair | 59,551,506 / 99.12% |
| Mapped reads, singletons | 227,800 / 0.38% |
| Secondary alignments | 0 |
| Supplementary alignments | 192,234 / 0.32% |
| Read min/max/mean length | 30 / 101 / 101.13 |
| Overlapping read pairs | 4,686,157 / 15.6% |
| Duplicated reads (flagged) | 0 / 0% |
| Duplicated reads (estimated) | 20,731,541 / 34.51% |
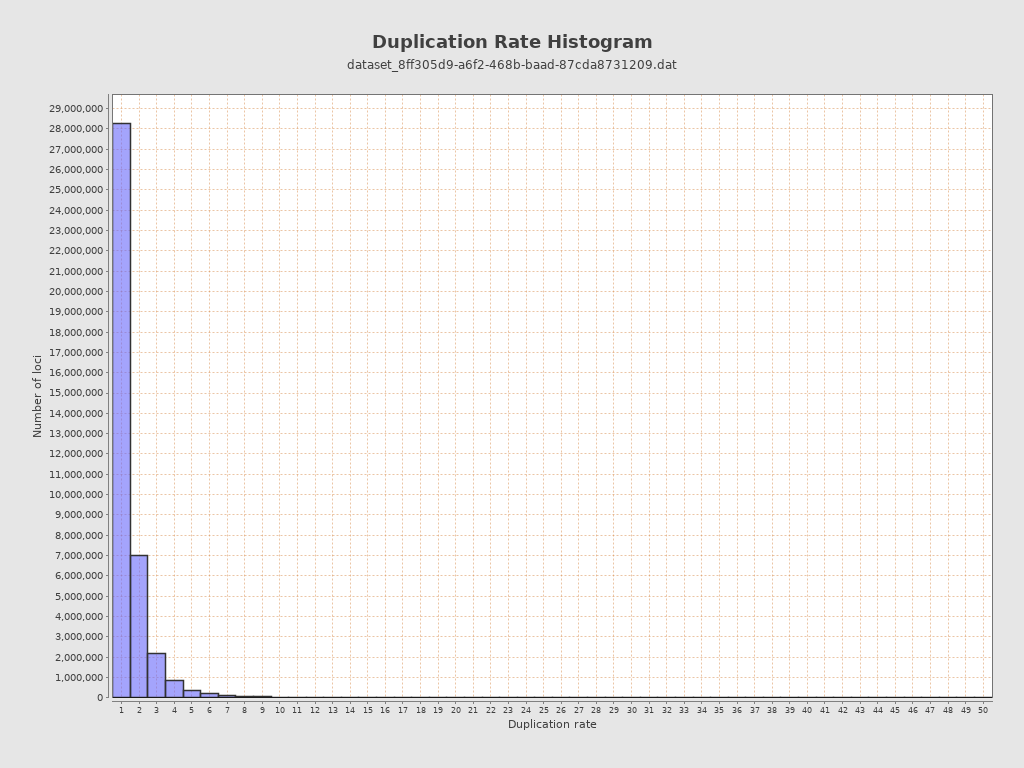
| Duplication rate | 28% |
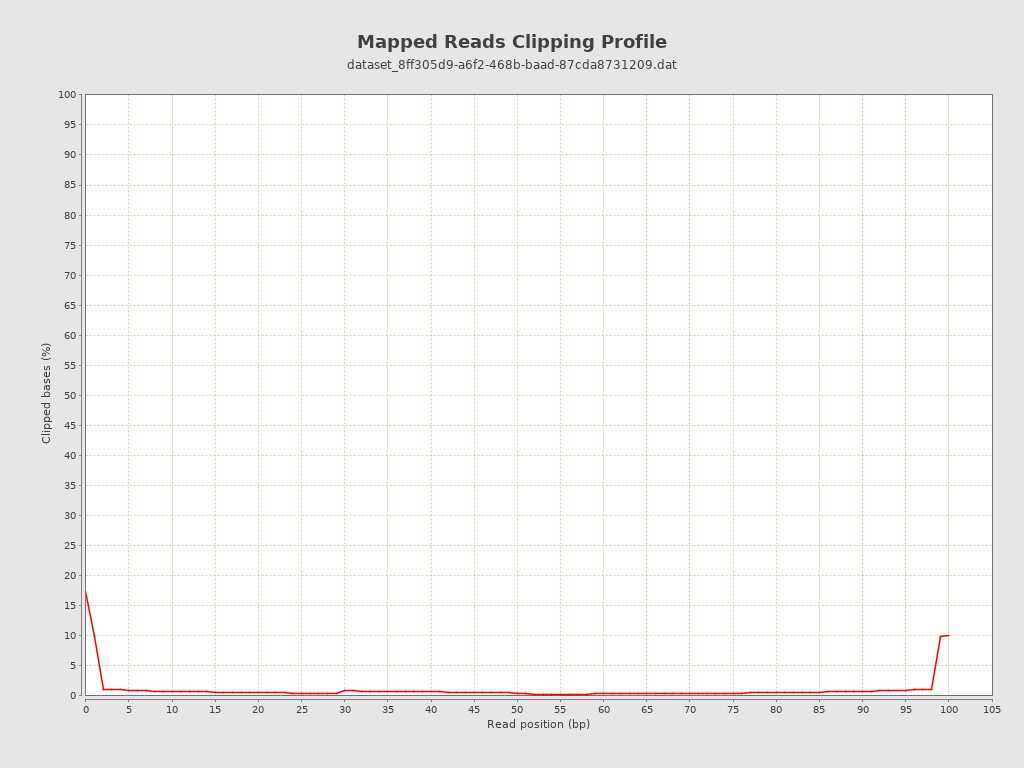
| Clipped reads | 15,202,268 / 25.3% |
ACGT Content
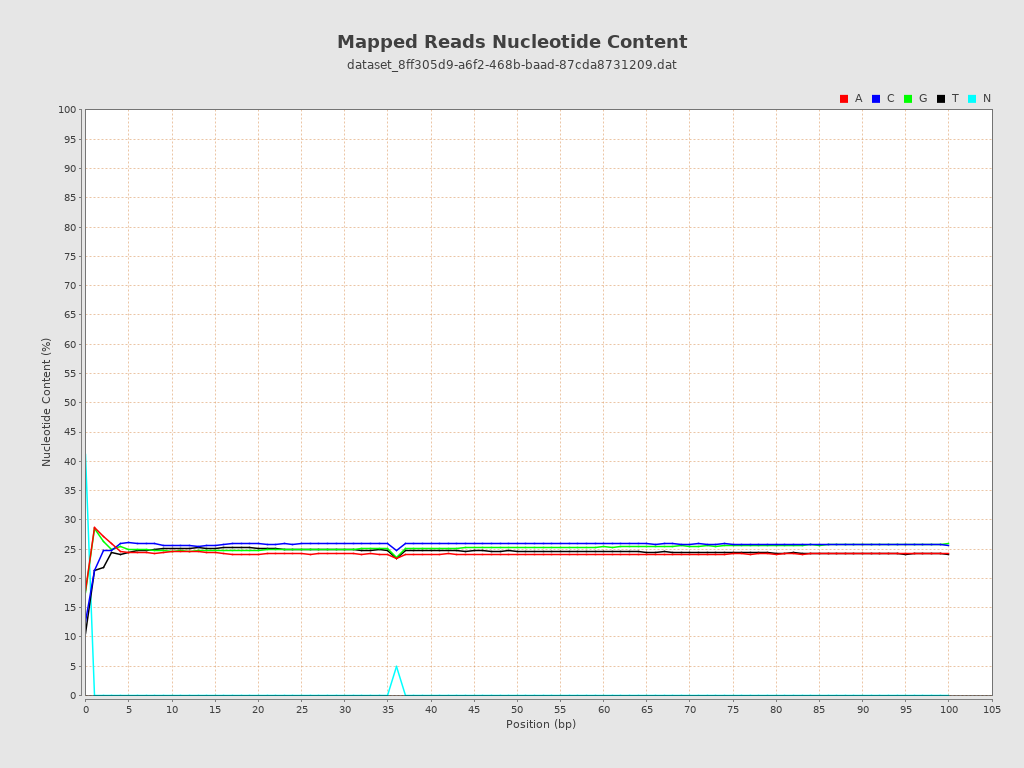
| Number/percentage of A's | 1,448,719,488 / 24.23% |
| Number/percentage of C's | 1,535,475,323 / 25.68% |
| Number/percentage of T's | 1,461,235,993 / 24.44% |
| Number/percentage of G's | 1,508,627,649 / 25.23% |
| Number/percentage of N's | 24,674,837 / 0.41% |
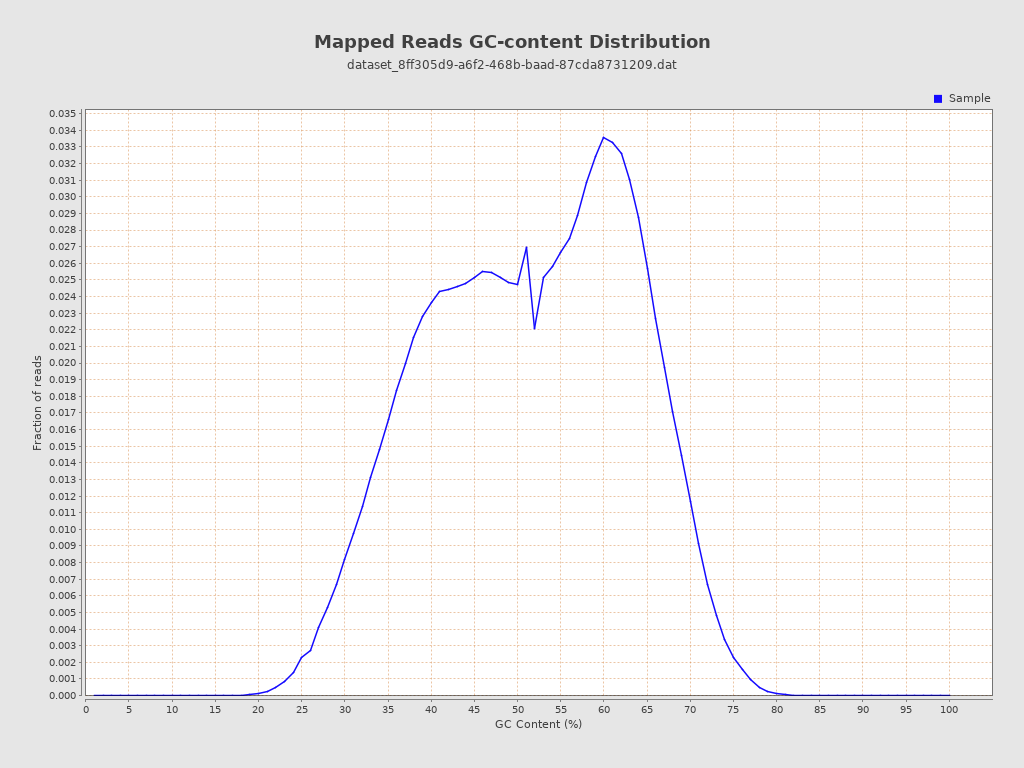
| GC Percentage | 50.92% |
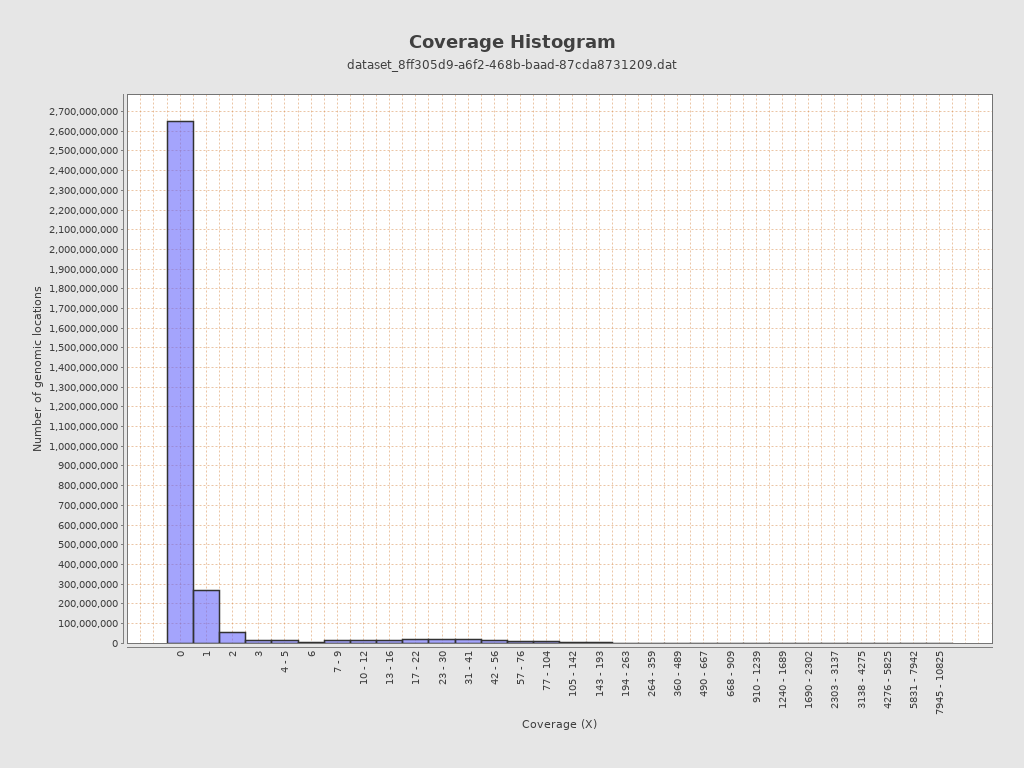
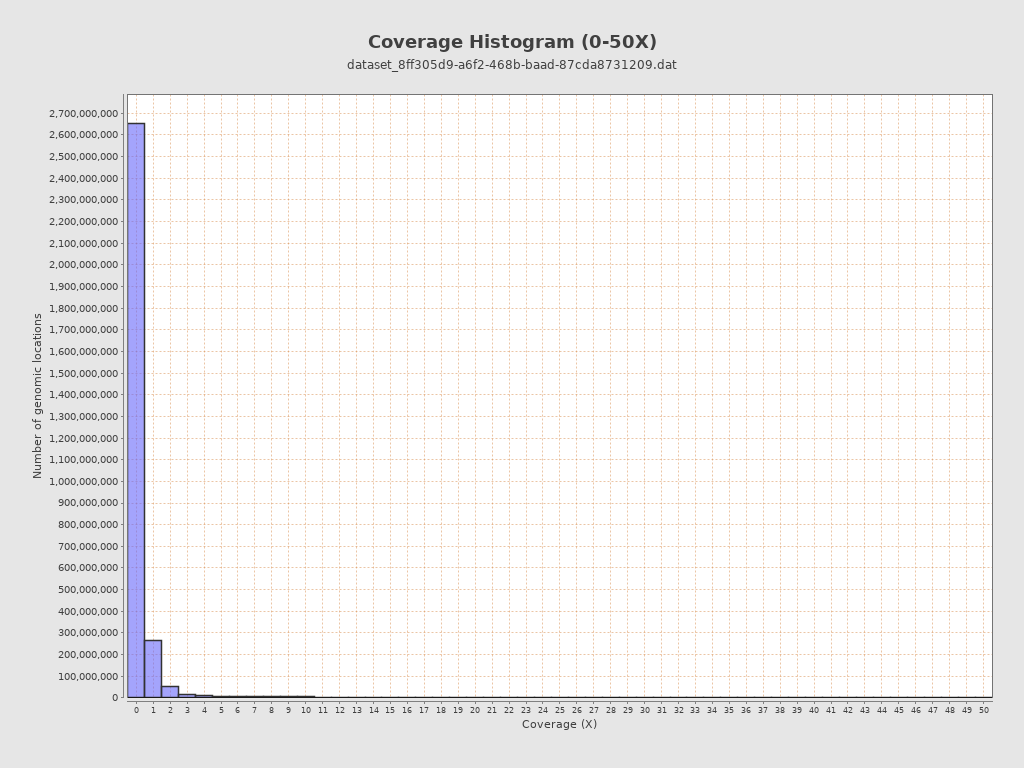
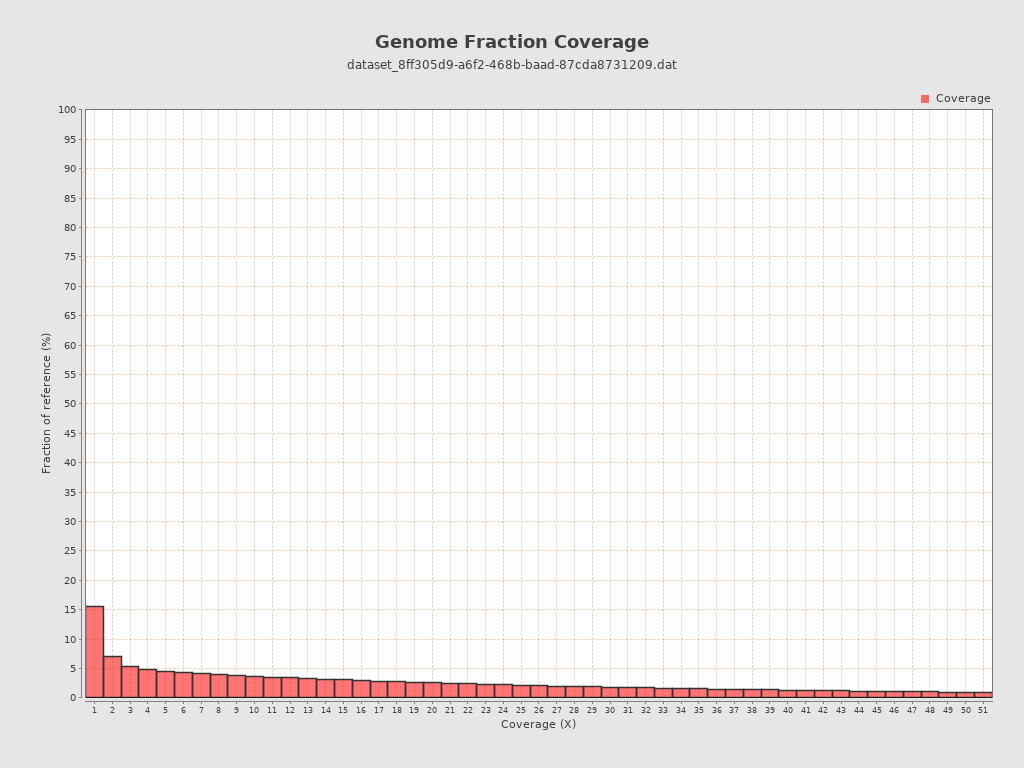
Coverage
| Mean | 1.906 |
| Standard Deviation | 18.6253 |
| Mean (paired-end reads overlap ignored) | 1.86 |
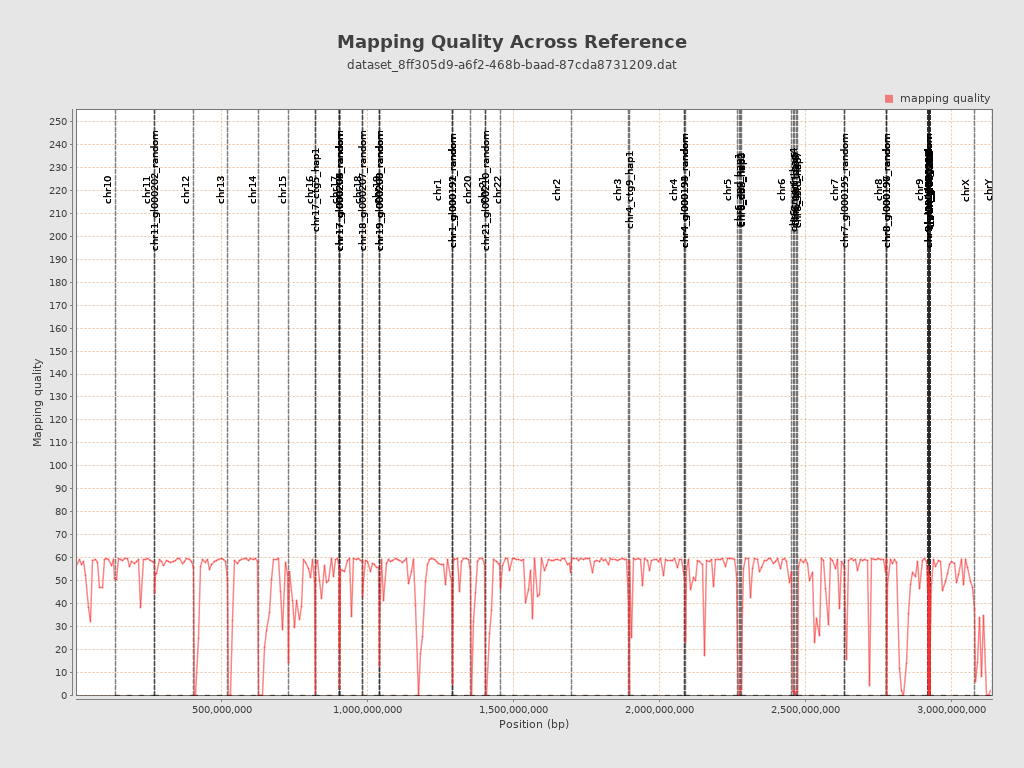
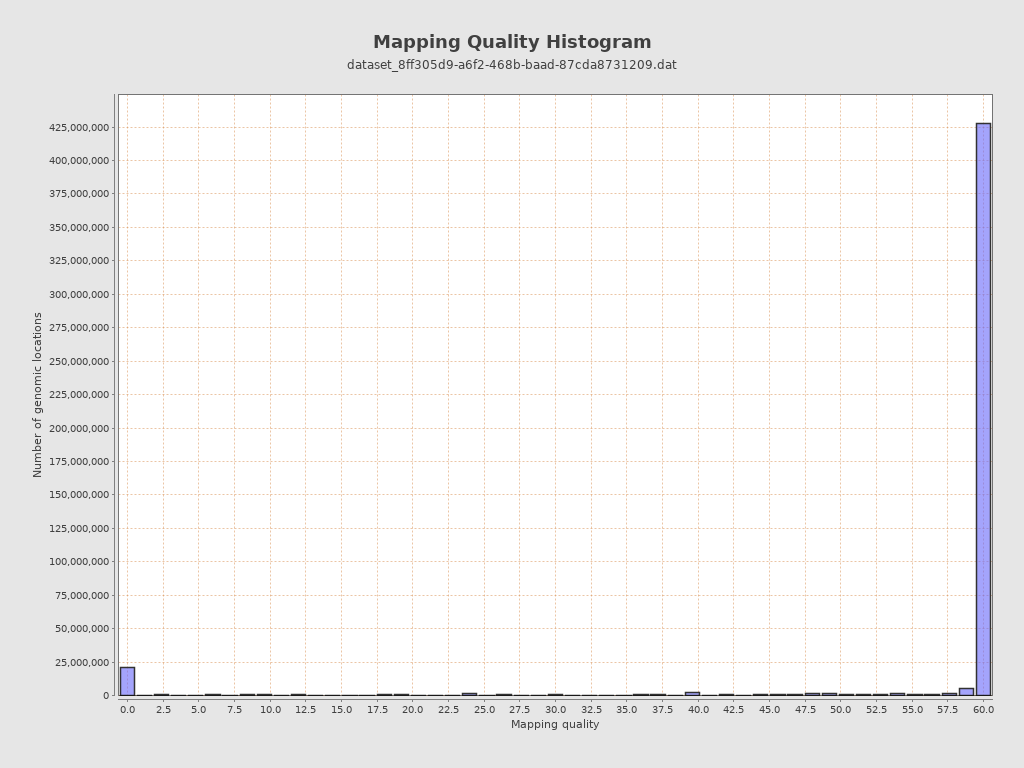
Mapping Quality
| Mean Mapping Quality | 46.46 |
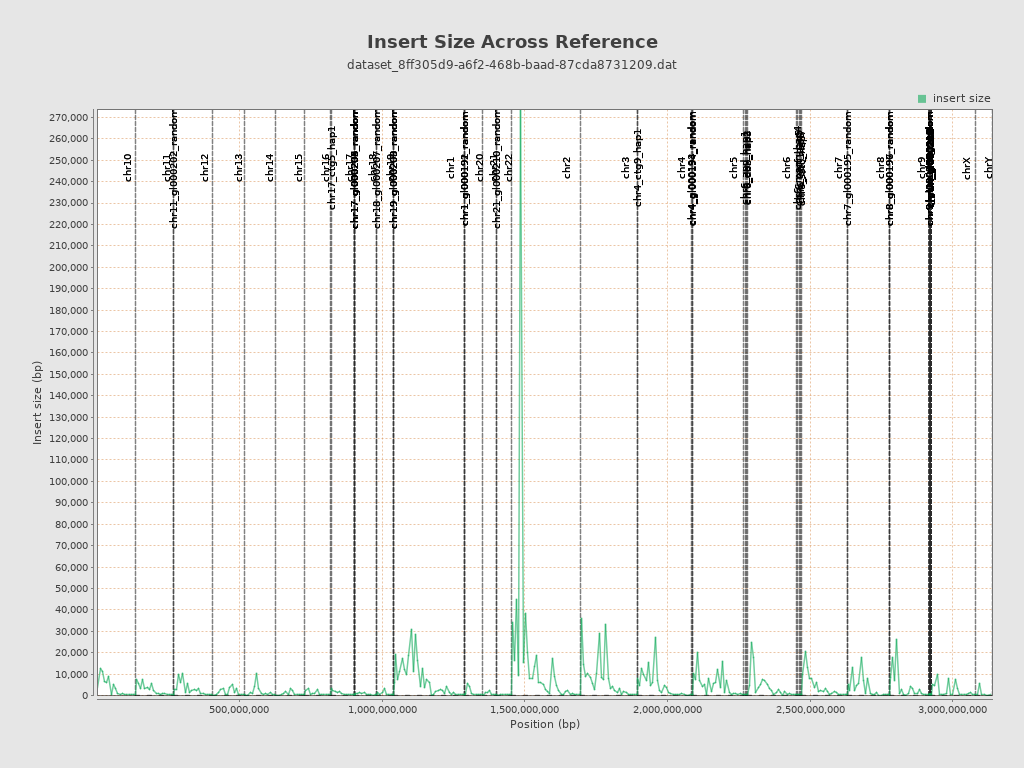
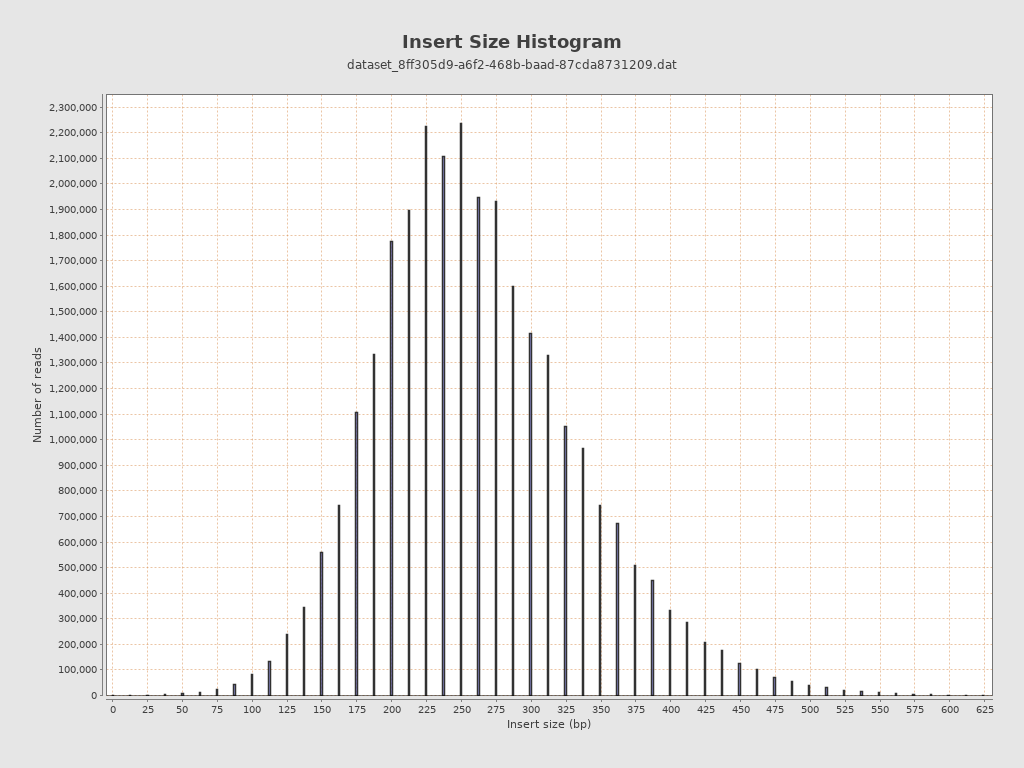
Insert size
| Mean | 87,640.6 |
| Standard Deviation | 2,932,065.54 |
| P25/Median/P75 | 218 / 260 / 312 |
Mismatches and indels
| General error rate | 0.7% |
| Mismatches | 41,170,236 |
| Insertions | 248,455 |
| Mapped reads with at least one insertion | 0.41% |
| Deletions | 325,235 |
| Mapped reads with at least one deletion | 0.54% |
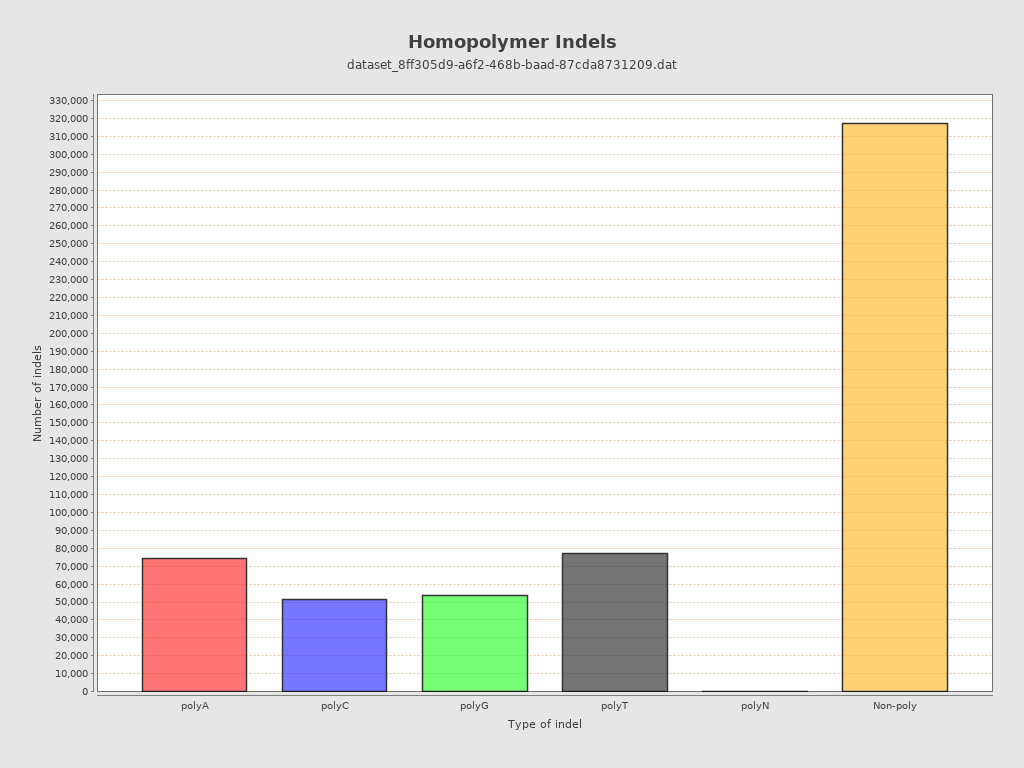
| Homopolymer indels | 44.7% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| chr10 | 135534747 | 234628752 | 1.7311 | 12.6836 |
| chr11 | 135006516 | 303132866 | 2.2453 | 13.5668 |
| chr11_gl000202_random | 40103 | 5797 | 0.1446 | 0.4689 |
| chr12 | 133851895 | 271920550 | 2.0315 | 12.3128 |
| chr13 | 115169878 | 103953309 | 0.9026 | 8.0372 |
| chr14 | 107349540 | 161743671 | 1.5067 | 12.7155 |
| chr15 | 102531392 | 262315063 | 2.5584 | 17.3908 |
| chr16 | 90354753 | 256719769 | 2.8412 | 16.9853 |
| chr17_ctg5_hap1 | 1680828 | 3156063 | 1.8777 | 10.8076 |
| chr17 | 81195210 | 329468385 | 4.0577 | 20.8248 |
| chr17_gl000203_random | 37498 | 4672 | 0.1246 | 0.3516 |
| chr17_gl000204_random | 81310 | 20429 | 0.2512 | 1.4427 |
| chr17_gl000205_random | 174588 | 1078809 | 6.1792 | 50.7138 |
| chr17_gl000206_random | 41001 | 6112 | 0.1491 | 0.5247 |
| chr18 | 78077248 | 92627126 | 1.1864 | 13.6508 |
| chr18_gl000207_random | 4262 | 2750 | 0.6452 | 1.2418 |
| chr19 | 59128983 | 311555587 | 5.2691 | 21.4877 |
| chr19_gl000208_random | 92689 | 72278 | 0.7798 | 2.4545 |
| chr19_gl000209_random | 159169 | 358143 | 2.2501 | 9.5238 |
| chr1 | 249250621 | 621794437 | 2.4947 | 19.9281 |
| chr1_gl000191_random | 106433 | 154112 | 1.448 | 5.4248 |
| chr1_gl000192_random | 547496 | 1368824 | 2.5002 | 10.0975 |
| chr20 | 63025520 | 131885964 | 2.0926 | 13.6925 |
| chr21 | 48129895 | 67378574 | 1.3999 | 14.0423 |
| chr21_gl000210_random | 27682 | 4564 | 0.1649 | 0.5101 |
| chr22 | 51304566 | 151623177 | 2.9554 | 20.9952 |
| chr2 | 243199373 | 388127937 | 1.5959 | 12.4924 |
| chr3 | 198022430 | 307879715 | 1.5548 | 12.2034 |
| chr4_ctg9_hap1 | 590426 | 614241 | 1.0403 | 6.6418 |
| chr4 | 191154276 | 227568430 | 1.1905 | 14.4987 |
| chr4_gl000193_random | 189789 | 375901 | 1.9806 | 23.1464 |
| chr4_gl000194_random | 191469 | 1355376 | 7.0788 | 61.3445 |
| chr5 | 180915260 | 296569604 | 1.6393 | 18.8765 |
| chr6_apd_hap1 | 4622290 | 2617632 | 0.5663 | 4.9216 |
| chr6_cox_hap2 | 4795371 | 6312152 | 1.3163 | 6.1029 |
| chr6_dbb_hap3 | 4610396 | 6487797 | 1.4072 | 6.6999 |
| chr6 | 171115067 | 239918287 | 1.4021 | 10.4455 |
| chr6_mann_hap4 | 4683263 | 4011793 | 0.8566 | 3.6696 |
| chr6_mcf_hap5 | 4833398 | 5847769 | 1.2099 | 5.2487 |
| chr6_qbl_hap6 | 4611984 | 5481256 | 1.1885 | 5.5674 |
| chr6_ssto_hap7 | 4928567 | 5034299 | 1.0215 | 4.7666 |
| chr7 | 159138663 | 371761424 | 2.3361 | 30.2983 |
| chr7_gl000195_random | 182896 | 994962 | 5.44 | 47.9125 |
| chr8 | 146364022 | 334126279 | 2.2828 | 46.5198 |
| chr8_gl000196_random | 38914 | 17686 | 0.4545 | 2.0136 |
| chr8_gl000197_random | 37175 | 31904 | 0.8582 | 3.9145 |
| chr9 | 141213431 | 269179800 | 1.9062 | 15.4447 |
| chr9_gl000198_random | 90085 | 127806 | 1.4187 | 11.5954 |
| chr9_gl000199_random | 169874 | 677280 | 3.987 | 10.9386 |
| chr9_gl000200_random | 187035 | 139429 | 0.7455 | 5.1652 |
| chr9_gl000201_random | 36148 | 88567 | 2.4501 | 6.0566 |
| chrM | 16571 | 258122 | 15.5767 | 4.7585 |
| chrUn_gl000211 | 166566 | 1256124 | 7.5413 | 21.1364 |
| chrUn_gl000212 | 186858 | 2664097 | 14.2573 | 65.0181 |
| chrUn_gl000213 | 164239 | 839267 | 5.11 | 43.8417 |
| chrUn_gl000214 | 137718 | 2375591 | 17.2497 | 50.3302 |
| chrUn_gl000215 | 172545 | 422966 | 2.4513 | 12.975 |
| chrUn_gl000216 | 172294 | 315091 | 1.8288 | 9.4388 |
| chrUn_gl000217 | 172149 | 279967 | 1.6263 | 12.6588 |
| chrUn_gl000218 | 161147 | 853658 | 5.2974 | 31.3587 |
| chrUn_gl000219 | 179198 | 765271 | 4.2705 | 41.1478 |
| chrUn_gl000220 | 161802 | 5775855 | 35.6971 | 250.4708 |
| chrUn_gl000221 | 155397 | 1664534 | 10.7115 | 44.0468 |
| chrUn_gl000222 | 186861 | 922246 | 4.9355 | 32.339 |
| chrUn_gl000223 | 180455 | 334780 | 1.8552 | 6.516 |
| chrUn_gl000224 | 179693 | 347018 | 1.9312 | 6.7492 |
| chrUn_gl000225 | 211173 | 749376 | 3.5486 | 25.3449 |
| chrUn_gl000226 | 15008 | 447807 | 29.8379 | 22.4501 |
| chrUn_gl000227 | 128374 | 598805 | 4.6645 | 27.7495 |
| chrUn_gl000228 | 129120 | 665638 | 5.1552 | 18.9052 |
| chrUn_gl000229 | 19913 | 351283 | 17.6409 | 44.5 |
| chrUn_gl000230 | 43691 | 54806 | 1.2544 | 4.5911 |
| chrUn_gl000231 | 27386 | 211747 | 7.7319 | 28.0881 |
| chrUn_gl000232 | 40652 | 88115 | 2.1675 | 12.513 |
| chrUn_gl000233 | 45941 | 3996 | 0.087 | 0.3239 |
| chrUn_gl000234 | 40531 | 126889 | 3.1307 | 21.4072 |
| chrUn_gl000235 | 34474 | 51080 | 1.4817 | 10.239 |
| chrUn_gl000236 | 41934 | 161688 | 3.8558 | 15.6967 |
| chrUn_gl000237 | 45867 | 198034 | 4.3176 | 22.8456 |
| chrUn_gl000238 | 39939 | 292633 | 7.327 | 22.1315 |
| chrUn_gl000239 | 33824 | 63424 | 1.8751 | 7.0195 |
| chrUn_gl000240 | 41933 | 29545 | 0.7046 | 3.8214 |
| chrUn_gl000241 | 42152 | 204988 | 4.8631 | 18.1401 |
| chrUn_gl000242 | 43523 | 220235 | 5.0602 | 16.1316 |
| chrUn_gl000243 | 43341 | 61582 | 1.4209 | 7.9595 |
| chrUn_gl000244 | 39929 | 194042 | 4.8597 | 30.7823 |
| chrUn_gl000245 | 36651 | 9539 | 0.2603 | 0.5533 |
| chrUn_gl000246 | 38154 | 20822 | 0.5457 | 3.3715 |
| chrUn_gl000247 | 36422 | 8944 | 0.2456 | 1.1009 |
| chrUn_gl000248 | 39786 | 5542 | 0.1393 | 0.4235 |
| chrUn_gl000249 | 38502 | 44644 | 1.1595 | 4.3416 |
| chrX | 155270560 | 134342701 | 0.8652 | 6.7699 |
| chrY | 59373566 | 38876606 | 0.6548 | 10.4863 |